Ewert Bengtsson, Patrik Malm, Hyun-Ju Choi, Bo Nordin
Partners: Rajesh Kumar, CDAC, Centre for Development of Advanced Computing, Thiruvananthapuram, India and K Sujathan, Regional Cancer Centre, Thiruvananthapuram, India.
Funding: Swedish Governmental Agency for Innovation Systems (Vinnova) and Swedish Research Council
Period: 0801-
Abstract: Cervical cancer is killing a quarter million women every year. Screening based on so called PAP-smears have proven very effective to reduce cancer mortality but require much work of well trained cytotechnologist. For 50 years research to automate the screening has been in progress, Bengtsson was very active in this field 1973-1993. Since about 10 years, commercial automated systems have been in operation but unfortunately those systems have many limitations. In India there is no effective screening program in operation and around 70,000 women die from the disease each year. Now an effort to develop a screening system adapted to Indian situation is in progress at the research institute CDAC in Thiruvananthapuram, Kerala in cooperation with the RCC, Regional Cancer Centre there. Based on our earlier experience in the field we are cooperating with this project and we have received support from Vinnova and VR for this. At CDAC they have developed a program for data collection which so far has been used at RCC to collect image data from around 800 patients. They have also developed an overall framework for the screening including image segmentation, artifact rejection, feature extraction and cell and specimen classification. They are currently evaluating the performance and working on improving the various parts of the system as needed. We are in particular studying whether the 3D chromatin texture of the cervical cells can be utilized as a robust feature for detecting (pre-) cancerous lesions. For that purpose we are scanning the cells at 40 different focus levels creating stacks of data for each cell nucleus. Dr Sujathan spent two weeks in Uppsala in October 2010 and another two weeks in October 2011 to collect image stacks and annotate cell nuclei with labels indicating the biological ``ground truth'' to be used for training and testing our classifiers. Dr Hyun-Ju Choi from Korea who was a post-doc at CBA 2008-2010 has studied 3D nuclear texture for other applications and we are currently evaluating her approaches for this purpose. In November 2011 we started collaboration with Andrew Mehnert at MedTech West, Chalmers through which the texture approaches he developed in his PhD thesis will be applied and evaluated. We have also implemented several other feature extraction methods and adapted them for our data stacks. Bengtsson spent the first week of 2011 in Thiruvananthapuram discussing in detail how our approaches could be integrated. These discussions continued in October 2011 when a delegation from CDAC visited CBA.
Martin Simonsson, Carolina Wählby
Partners: Johen Kreuger, Sara Thorslund, Gradientech AB, Uppsala
Funding: SciLife Lab Uppsala
Period: 1108-
Abstract: Tracking of cell movements in various cell culture setups is essential to many researchers in the life science sector. Gradientech AB, a Swedish biotech company, has developed CellDirector, a unique microfluidic system that academic researchers can use to study how concentration gradients of soluble proteins impact cell migration. The current project is focused on developing software for analyzing cell behavior and cell migration. The free open-source software CellProfiler developed at the Broad Institute will be used as a platform for a high-throughput system with automated high quality imaging, adapted for unlabeled cells, which are analyzed with regard to directionality of migration, speed, and acceleration. Apart from analyzing cell migration, the cell tracking aims at producing lineages, where cellular events such as cell division and cell death can be scored for single cells.
Carolina Wählby
Partners: Jeremie Zoueu, Olivier Bagui, Dept. Genie Electrique et Electronique, Institut National Polytechnique, Felix Houbhouet-Boigny, Cote d' Ivoire
Period: 1109-
Abstract: This project aims to propose an effective optical device based on LED spectral microscopy, which will be low cost, fast and easy to use in the diagnosis of human malaria parasites, especially because the sample will not need any special preparation or staining and the data will be automatically processed to provide real-time diagnosis of the type of the parasite, the parasitic density and its age for an effective prescription. The collaborative project was initiated by a 3-month visit by Olivier Bagui, where we focused on the development of efficient segmentation methods for unstained images of blood cells.
Ida-Maria Sintorn, Robin Strand
Partners: Ingela Parmryd, Dept. of Medical Cell Biology, UU; Jeremy Adler, Dept. Of Immunology, Genetics and Pathology, UU
Funding: TN-faculty UU; S-faculty, SLU; VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 1101-
Abstract: A cell surface is a highly irregular and rough. The surface topography is however usually ignored in current models of the plasma membrane, which are based on 2D observations of diffusion that really occurs in 3D. In this project we model diffusion on non-flat surfaces to explain biological processes occurring on the cellsurface.
Bettina Selig, Cris Luengo
Partners: Bernd Rieger, Quantitative Imaging Group, Delft University of Technology, Netherlands; Koen Vermeer, Eye Hospital Rotterdam, Netherlands
Funding: S-faculty, SLU
Period: 1103-
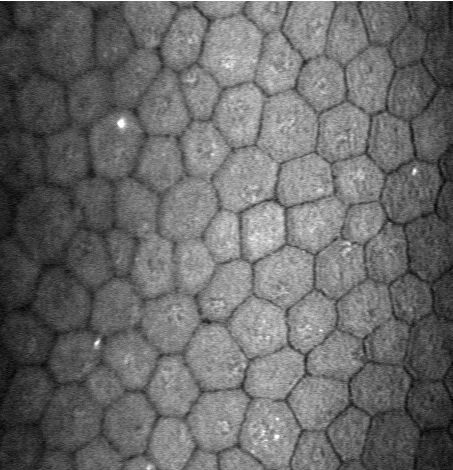
Abstract: In many corneal studies, endothelial cell density and morphology is used to assess the quality of the cornea. Based on these parameters, important therapeutic decisions are made. The endothelium may be imaged by specular microscopy or by confocal scanners and measurements can be obtained manually, automatically with manual corrections or fully automatically with current software (e.g., Nidek's Navis). Unfortunately, the results of the automatic mode are insufficient (see Figure 8) when the image quality is affected or when irregular shaped endothelial cells are present. In this project, we are developing a new segmentation method, using stochastic watershed, that enables a better estimation of endothelial quality.


|
Milan Gavrilovic, Carolina Wählby
Partners: Irene Weibrecht, Tim Conze, Ola Söderberg, Ulf Landegren, Dept. of Genetics and Pathology, UU
Funding: TN-faculty, UU
Period: 0807-
Abstract: Our previously published novel methods for quantification of colocalization are applied to a number of projects in collaboration with the Molecular Tools group at the Dept. of Genetics and Pathology. The aim is detection of multi-color rolling circle products representing DNA-protein interactions in wide-field fluorescence microscopy images, as recently published (Weibrwcht et al., New Biotechnology). In addition, we use the methods for evaluation of biochemical methods relevant in early stages of specimen preparation, e.g., estimating the quality of oligonucleotides, as presented in (Gavrilovic et al., Cytometry).
Amin Allalou, Carolina Wählby
Partners: Carlos Pardo, Mehmet F. Yanik, Research Laboratory of Electronics,
Massachusetts Institute of Technology, Cambridge, USA
Funding: TN-faculty, UU, The Swedish Research Council, Collaboration Grant, Medicine
Period: 1009-
Abstract: The research collaboration initiated during 2010 continued with more visits to MIT, Cambridge, USA, during 2011. A high-throughput platform for cellular resolution in vivo chemical and genetic screens on zebrafish has been developed at MIT, Cambridge, USA. The system automatically loads zebrafish larvae and positions and rotates them for high-speed confocal imaging. The number of neurons in the fish is of importance to different screens and therefore a method has been tested to count the number of neurons in certain regions of the zebrafish brain. In addition, some methods have been developed to rotate and align the fish correctly before imaging, as published in (Chang et al., Lab. Chip). The automated rotational positioning also inspired the development of optical computer tomography methods for accurate and precise 3D volume rendering of zebrafish embryos, currently under development.
Amin Allalou, Ida-Maria Sintorn, Carolina Wählby
Partners: Carl-Magnus Clausson, Ola Söderberg, Dept. of Genetics and Pathology, UU
Funding: TN-faculty, UU; VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 1002-
Abstract: A novel approach to increase the dynamic range of in situ PLA has been developed at Dept. of Genetics and Pathology, UU. Using several probes with different concentrations the dynamic range can be extended significantly. Signal detection previously developed at CBA, UU, (3DSWD) is used to quantify the number of signals in the different concentrations (Clausson et. al. Nature Methods).
Khalid Niazi, Ewert Bengtsson
Partners: Mats Nilsson, Lennart Dencker, Div. of Toxicology, Dept. of Pharmaceutical Biosciences, UU; Bill Webster, School of Medical Sciences, University of Sydney, Australia
Funding: Swedish Research Council with Lennart Dencker as PI, COMSATS IIT, Islamabad
Period: 0711-1111
Abstract: Embryo cultures of rodents is an established technique for monitoring adverse effects of chemicals on embryonic development. The cultures are assessed both morphologically looking for changes in the development of various organ structures and through analysis of the heart rate of the embryo where irregularities are a sign of adverse influences of the tested drug.
The heart rate was determined through fully automated image analysis of video sequences. After finding the heart location in the image through spatio-temporal image analysis we modeled the movement of the heart as a sinusoid which was analyzed using Emperical Mode Decomposition (EMD). We used EMD with slight modification along with Laplacian Eigenmaps to detect the periodic activity. The normal embryo's heart activity can easily be detected by local maxima detection but it becomes a challenging task once the heart activity becomes abnormal. The developed methods functions well and are being used by our pharmacological partners.
We also studied how the currently used subjective, semi-quantitative (categorical, score) assessment of the morphological development could be made more objective and quantitative. We initially created automatically extracted descriptors for the different organs that were to be related to the subjective assessments. The outcome of this needs to be evaluated further.
This project was presented as part of the PhD thesis by Khalid Niazi that was defended in November 2011.
Khalid Niazi, Bettina Selig, Ewert Bengtsson, Ingela Nyström
Partner: Albert Alm, Dept. of Neurosciences, UU
Funding: COMSATS IIT, Islamabad
Period: 0711-
Abstract: Retinal imaging is one of the main sources in ophthalmology to study the optical nerve head and the retina. Retinal images are often used for analyzing, diagnosing and treating a number of diseases of the human retina. Image registration plays an important role in determining the progression of retinal illness. In the current project, we are developing a method which will help in evaluation of glaucoma progression. We are especially concentrating on correction of parallax error, which is normally produced due to a change in the angular position of the camera.
Retinal image registration can be performed between either the full images or within sub-regions. The movement of vessels makes it illogical to perform registration between full images. Using a sub-region which is least effected by vessel movement will present a true picture of the vessel movement. The vessels inside the optic disc, which lie close to the origin, move with time and often get over-exposed during imaging. It has also been reported that the end of the vessel gets detached from the surface of the retina due to loss of the nerve fibers, which leaves us to use the area around the border of the optic disc. We have used the conventional particle swarm optimization (PSO) algorithm which uses uniform distribution to update the velocity equation. Subsequently, we have modified the PSO algorithm which utilizes benefits from the Gaussian and the uniform distribution, when updating the velocity equation. Which one of the distributions is selected depends on the direction o f the cognitive and social components in the velocity equation. This direction checking and selection of the appropriate distribution provide the particles with an ability to jump out of local minima. The registration results achieved by this new version proves the robustness and its ability to find a global minimum.
This project was presented as part of the PhD thesis by Khalid Niazi that was defended in November 2011.
Ingrid Carlbom, Milan Gavrilovic, Ewert Bengtsson, Jimmy Azar
Partners: Christer Busch, Marene Landström, Dept. of Genetics and Pathology, UU Hospital
Funding: Swedish Research Council
Period: 1001-
Abstract: Prostate cancer diagnosis is based on Gleason grading, which is the most widely used system for determining the severity of prostate cancer from tissue samples. However, Gleason grading is highly subjective with significant variation between experienced pathologists, which studies show may be as high as 30-40%. We propose to replace subjective diagnosis of prostate cancer with automatic severity grading using a combination of tissue staining and image analysis. The goal of this research is to identify and separate slow-growing cancer from more aggressive types, thereby helping to reduce needless radical treatment of the disease. Currently about 70% of patients with localized prostate cancer receive aggressive treatment that does not prolong life but often results in debilitating side effects.
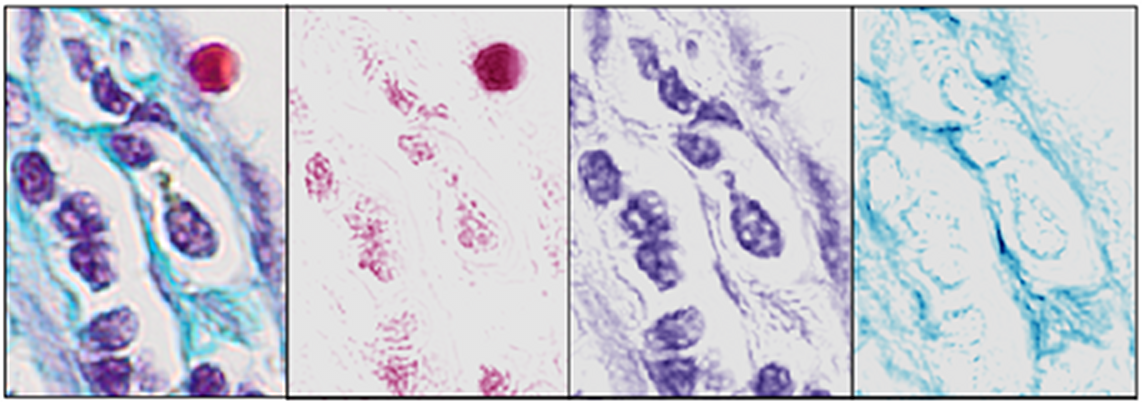
We have developed an automatic (blind) method, referred to as the BCD (Blind Color Decomposition) method, for color decomposition of histological images acquired with a RGB camera. The method decouples intensity from color information and bases the decomposition only on the tissue absorption characteristics of a specific stain. When a stain does not absorb light but rather scatters light, we automatically remove the areas affected by that stain from the image prior to color decomposition. By modeling the CCD sensor noise with statistical techniques, we improve the method's accuracy. We extend current linear decomposition methods to include stained tissues where one of their spectral signatures cannot be separated from all combinations of the other tissues' spectral signatures. The result of the method is a set of density maps, one for each stained tissue type, which classifies portions of pixels into the correct stained tissue thereby reducing aliasing artifacts (See Figure 9). Comparisons with current color decomposition methods demonstrate that our method outperforms methods in the literature, giving a 92% median correlation with ground truth as compared to other published methods that give up to 81% median correlation. The BCD method produces highly accurate density maps that can be used for identification of morphological features that are linked to cancer. This work resulted in a US provisional patent application.
During 2010, we developed a new staining method combining histochemical and immunohistochemical stains, making it possible to discriminate between normal glandular structures and infiltrating cancer. We have continued to develop stains that also give a color decomposition into density maps which enable highly accurate measurements of morphological features for determining the malignancy grade. We are in the process of evaluating several stains using methods for quantitative determination of the efficacy of stain/tissue combinations.
We developed a simple, computationally efficient, automated method for accurate detection and localization of cores in tissue microarray images (TMAs), which is based on geometric restoration of core shapes in microarray images without any assumptions on grid geometry. The method relies on hierarchical clustering in conjunction with the Davies-Bouldin index for cluster validation in order to estimate the number of cores present in the image wherefrom we estimate the core radius and refine this estimate using morphological granulometry. The final stage of the algorithm reconstructs circular discs from core sections such that these discs cover the entire region of the core regardless of the precise shape of the core. The method was tested on over 32 TMA images comprising over 2300 cores. The results show that the proposed method is able to reconstruct the location of the cores without any evidence of localization error even if only a partial core is included in the slide. Furthermore, an assessment of the computational efficiency of the algorithm shows it to be far more efficient than existing methods based on the Hough transform for circle detection.
Amin Allalou, Carolina Wählby
Partners: Nils-Göran Larsson, Dept. of Laboratory Medicine, Karolinska Institute; Mats Nilsson, Chatarina Larsson, Dept. of Genetics and Pathology, UU
Funding: The Swedish Research Council, Collaboration Grant, Medicine
Period: 0801-
Abstract: Mutations of mitochondrial DNA (mtDNA) cause genetic syndromes with widely varying phenotypes and are also implicated in many age-associated diseases and the ageing process itself. Our knowledge of the principles governing segregation of mtDNA mutations in somatic tissues and in the germ line is very limited. In this collaborative project we combine a powerful technique for detection of individual mtDNA molecules with image analysis. We work with a variety of mouse models and the goal is to develop image analysis software to do three-dimensional (3D) reconstruction of the distribution of mutated mtDNA molecules in mammalian tissues. We want to use this technology to study segregation of mtDNA mutations in mouse tissues and to study the mtDNA bottleneck by visualizing the distribution of mutated mtDNA during oogenesis. The ultimate goal is to study the distribution of mtDNA mutations in embryos and placenta to establish principles for prenatal diagnosis.
Ida-Maria Sintorn
Partners: Adrian Baddeley, Michael Buckley, Leanne Bischof, CSIRO Mathematical and Information Sciences, Australia; Stephen Haggarty, Broad Institute of Harvard and MIT, USA
Funding: S-faculty, SLU; VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 0901-
Abstract: In biological research and in the drug development process when screening for new drugs, HCA systems are often used. Such a system is a fully automatized microscopy system that acquires hundreds or thousands of images in an experiment and automatically extracts information from the image data. In this project we have developed pre-processing tools to allow for improved comparison of image content within and across cellular screening experiments. We also develop methods and new statistical analysis tools for so called co-culture HCA experiments i.e., when more than one celltype are cultured together to allow for investigating their interaction in response to added substrates. During 2011 Sintorn spent three weeks at the Broad Institute as a guest researcher working on this project.
Ida-Maria Sintorn
Partner: Vironova AB, Stockholm
Funding: VINNMER programme, Swedish Governmental Agency for Innovation Systems
Period: 0801-
Abstract: Electron Microscopy allows for studying the shape and morphology of biological particles such as viruses at the nm level. This means for example that structural differences between virus maturation stages, related virus species, wild type virus and virus treated with a potential drug or a small molecule can be analyzed. Both external (shape and protein patterns on the virus surface) and internal structural differences can be analyzed. In this project methods for efficiently identifying and quantifying such structural differences are developed.
Partners: Vironova AB, Stockholm; Delong Instruments, Brno, Czech Republic; Ali Mirazimi, Kjell-Olof Höglund, Swedish Institute for Communicable Disease Control (SMI); Jan-Olof Strömberg and Joel Andersson, Dept. of Mathematics, KTH
Funding: 2008-2011 Swedish Civil Contingencies Agency (MSB), Swedish Defense Materiel Administration (FMV), Swedish Agency for Innovative Systems (VINNOVA). 2011- Eurostar project E!6143.
Period: 0801-
Abstract: This project aims at automating the virus identification process in high resolution TEM images. This, in combination with project 28 create a rapid, objective, and user independent virus diagnostic system. The identification task consists of method development for segmenting virus particles with different shapes and sizes and extracting descriptive features of both shape and texture to enable the classification into virus species. Texture features such as variants of Local Binary Patterns are being evaluated on virus textures as well as other texture datasets to get a deeper understanding of the discriminant power of the features under different conditions. Figure 10 shows the texture of 15 virus types.
A paper evaluating the segmentation method was accepted to Journal of Microscopy in 2011. Work on texture features was presented at the 16th Iberoamerican Congress on Pattern Recognition (CIARP) 2011 in Pucón, Chile.
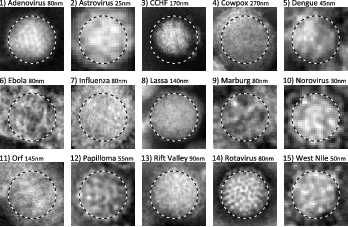
|
Partners: Vironova AB, Stockholm; Delong Instruments, Brno, Czech Republic; Ali Mirazimi, Kjell-Olof Höglund, Swedish Institute for Infectious Communicable Control (SMI); Gun Frisk and Monika Hodik, Dept. of Immunology, Genetics and Pathology, UU
Funding: 2008-2011 Swedish Civil Contingencies Agency (MSB), Swedish Defense Materiel Administration (FMV), Swedish Agency for Innovative Systems (VINNOVA). 2011- Eurostar project E!6143
Period: 0801-
Abstract: Transmission electron microscopy (TEM) is an important virus diagnostic tool. The main drawback is that an expert in virus appearance in electron microscopy needs to perform the analysis at the microscope, an often very time consuming task.
The project aim is to develop methods for a multi-scale analysis at the microscope to automatically acquire highly magnified images of possible virus particles. This is an important step towards automating the virus identification process and thereby creating a rapid, objective, and user independent virus diagnostic system. By introducing the multi-scale approach the search area where highly magnified images need to be acquired is estimated to be reduced with more than 99.99%.
As of mid 2011 Delong Instruments has joined the project. They will develop a novel bench-top low-voltage TEM where the methods for automated acquisition will be incorporated. This work will intensify the development of methods for the automatic acquisition of images.
Azadeh Fakhrzadeh, Cris Luengo, Gunilla Borgefors
Partners: Ellinor Spörndly-Nees, Lena Holm, Dept. of Anatomy, Physiology and Biochemistry, SLU
Funding: SLU (KoN)
Period: 1009-
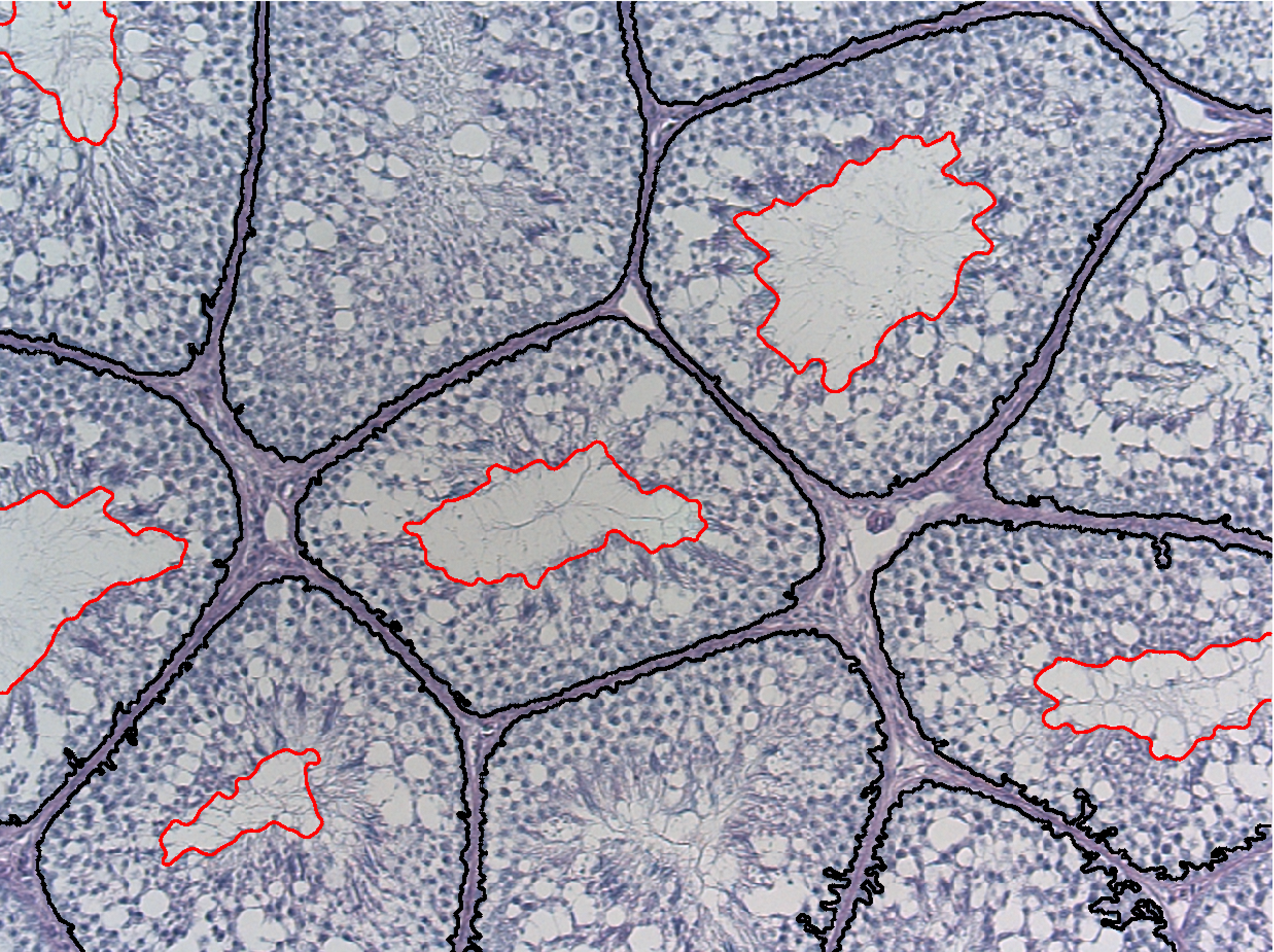
Abstract: Reproductive toxicology is the study of chemicals and their effects on the reproductive system of humans and animals. In reproductive toxicology, there is a strong need to detect structural differences in organs that often have both a complex microscopic structure and function. This problem is further complicated because standard techniques are based on the examination of two-dimensional sections of a three-dimensional structure. The aim of this project is to develop methods to objectively describe microscopic structures of male reproductive organs and to test these in reproductive toxicology research. The project is comparative and includes studies of organs from rooster and mink. We are developing automatic and interactive methods to analyze the relevant structures in the histology images of testis. We have constructed an automatic method to delineate the seminiferous tubule border and lumen, see Figure 11. We use a level set based active contour method to delineate the lumen border and classical classification scheme to detect the seminiferous border.
|
Hamid Sarve, Joakim Lindblad, Vladimir Curic, Gunilla Borgefors
Partners: Carina Johansson, Dept. of Clinical Medicine, Örebro University/Institute of Odontology, The Sahlgrenska Academy, Gteborg; Nataša Sladoje, Faculty of Technical Sciences, University of Novi Sad, Serbia
Funding: Swedish Research Council; S-faculty, SLU
Period: 0503-
Abstract: With an aging and increasingly osteoporotic population, bone implants are becoming more important to ensure the quality of life. In order to evaluate how tissue reacts on implants, the interface at the implant and tissue must be studied. Today, this is done manually in a microscope which is a costly and time-consuming procedure.
The aim of this project is to develop automatic image analysis methods for evaluating images of the interface region of tissue and implant. These methods would provide faster and more objective measurements on how well the implant is integrated in the bone compared to today's manual methods. The analysis involves segmentation of the images into different tissue-types and quantifying bone contact length, area and volume.
The project encompasses parallel development and comparison of methods for 2D analysis of histological sections as well as 3D analysis of SRCT volumes. Within the project, methods for segmentation and feature extraction have been developed for both 2D histological sections, and 3D SRCT volumes. To facilitate comparison of results from the two imaging modalities, a 2D-3D image registration method has also been developed. Furthermore, another significant contribution has been the development of methods for extraction of the 3D features from the 3D volume data. A thesis, titled Evaluation of Osseointegration using Image Analysis and Visualization of 2D and 3D Image Data, which was based on the methods described above were defended by Hamid Sarve in 2011.
Hamid Sarve, Joakim Lindblad, Gunilla Borgefors
Partners: Carina Johansson, Dept. of Clinical Medicine, Örebro UniversityDept. of Clinical Medicine, Örebro University/Institute of Odontology, The Sahlgrenska Academy, Gteborg; AstraTech, Mölndal
Funding: The Knowledge Foundation
Period: 0906-
Abstract: This project aims to develop new techniques for interactive 3D visualization of bone anchored implants in order to facilitate the understanding of the mechanisms of implant integration. To enable good communication between the people involved in development, production and use of medical devices - computer scientists, material scientists, and medical doctors - each with their own special knowledge, it is of highest importance to provide a common visual platform for a mutual understanding of the problems of implant integration. Being able to actually see the 3D structure around the implant for the first time will inspire new measures of the implant integration quality.
We base the visualization on data from non-destructive 3D SRCT imaging; this technique yields more accurate tomographic reconstruction at higher resolution compared to standard CT. Furthermore, SRCT-imaging is more suitable for samples containing metal, as the artefacts caused by the metal is significantly lower compared to traditional CT. Existing visualization software are not really useful for this type of complex and highly detailed data, requiring the development of special purpose methods and software.
The combination of this project and project 30 will improve both quantitative and qualitative analysis of bone implant integration and thereby support the development of more effective implants and diminish the number of malfunctioning devices.Two new methods for visualizing SRCT-scanned volume samples were presented at the International Conference on Computer Vision and Graphics (ICCVG'10); one being an animation that follows the implant thread and extracts information about the bone-implant integration over the whole sample and the other a 2D unfolding that displays a flattened version of the implant surface, with feature information projected onto it, providing a direct overview of the implant integration (see Figure 12). The methods were applied on real clinical data; they were applied in a case study involving retrieved human oral implants. As the case study showed, the use of 3D techniques highlighted the complexity of osseointegration and provided information other than the 2D analysis on histological images. The visualization techniques were further discussed in Hamid Sarve's thesis, Evaluation of Osseointegration using Image Analysis and Visualization of 2D and 3D Image Data, defended in 2011.

|
Andreas Kårsnäs, Robin Strand, Ewert Bengtsson
Partners: Visiopharm, Hørsholm, Denmark; Clinical Pathology Division, Vejle hospital, Vejle, Denmark
Funding: NordForsk Private Public Partnership PhD Programme and Visiopharm
Period: 0909-
Abstract: The results of analyses of tissue biopsies by pathologists are crucial for breast cancer patients. In particular, the precision of a patient's prognosis, and the ability to predict the consequences of various treatment opportunities before actually exposing the cancer patient, depend on the detection and quantification of biomarkers in tissue sections by microscopy. Experience from the last decade has revealed that manual detection and quantification of biomarkers by microscopy of tissue biopsies is highly dependent on the competencies and stamina of the individual pathologist. The aim of the present PhD project is to develop software-based algorithms that can facilitate the workflow and ensure objective and more precise results of the quantitative microscopy procedures in breast cancer.